How is the monitoring carried out?
A critical part of environmental monitoring is the widespread, accurate collection of water samples. The distribution of monitored species can change very rapidly, and the presence and abundance of species is often unknown in a particular area.
The collection of water samples is at the heart of this project. The engagement of citizen scientists has been important in monitoring both aquatic and terrestrial species. As part of this project, students and teachers help track the range of several invasive species throughout New York, which will greatly contribute to efforts to control the rapid spread of these destructive, costly pests. They also help track the locations of the endangered native species deepwater cisco and American eels, which maps habitat loss or gain.
There are a number of ways of monitoring a body of water for fish species, including visual sightings, catching or trapping, and, more recently, monitoring of species specific DNA from cells shed into the environment. We use a very sensitive technology that monitors DNA found in environmental samples (environmental DNA or eDNA). eDNA is genetic material that is found in environmental samples like water, soil, or air. eDNA can be either nuclear or mitochondrial DNA that is released into the environment as a result of the constant shedding of cells by all organisms into the environment. For example, fish cells can be shed into the water in multiple ways, including in mucous, feces, urine, or blood, or as flaked off skin cells. The cells shed into the environment all contain genetic material (DNA) that is unique to their species. When eDNA is collected, it is made up of DNA from all the different organisms present in the environment, including plants, animals, singled celled organism like protozoa, and bacteria.
Therefore, eDNA can be used to provide information about what organisms are or were recently present in a particular area.
When you isolate eDNA, you don’t know what DNA is in the sample until you conduct a genetic analysis. For example, if you collect a bucket of water from a lake and collect DNA from that water, you potentially have eDNA from all of the organisms living in that lake. If that lake has fish in it, that eDNA will contain some fish DNA, along with many other DNAs from unknown sources. eDNA monitoring has been used in both fresh water and marine environments and provides a highly efficient, sensitive, and cost-effective way to monitor for the presence of different species.
How is the eDNA collected?
We provide a kit containing all of the materials needed to collect and filter multiple water samples from each site to be tested. The water sample is pumped through porous filters that will retain any cells shed by fish in the collection area. Although there is free eDNA in the environment, for this test we will actually be looking at DNA contained in cells released into the environment. That’s because the filters we are using will generally not trap free DNA molecules, but will retain intact cells. Since the half life of DNA (the amount of time required for the amount of measurable DNA to fall to half its value as measured at the beginning of the time period) from fish cells shed into the fresh water is at least 4-6 hours following shedding, the presence of a fish can potentially be detected even if it swam by several hours earlier. The filter paper containing cells from the water sample is placed into a vial containing a solution that protects the DNA from further breakdown and is sent back to Cornell University for DNA extraction and qPCR analysis. A detailed description of the collection technique is available for download.
How is qPCR used to monitor for the presence of eDNA from different species?
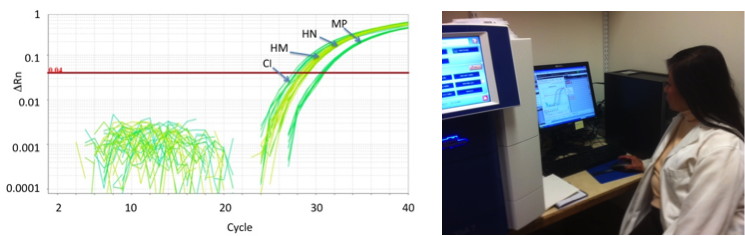
To determine if the water collected contained DNA from any of the species being monitored, total eDNA is extracted from the collection filter. A variation of the polymerase chain reaction (PCR) technique is used to determine if the eDNA sample contains DNA from any of the fish species being tested for. The polymerase chain reaction (PCR) is a way to make thousands or even millions of copies of a small, highly specific region of DNA, starting with a very tiny amount of DNA. Generally the copies of DNA made are analyzed at the end of the reaction, usually by running them on an agarose gel. Using standard PCR methods, only the final product made during the PCR reaction can be analyzed. Quantiative PCR (qPCR) is a variation of the standard PCR technique that allows an analysis of the DNA copies being made in real time, as the reaction is actually going on. During the qPCR process, a fluorescent dye is incorporated into the newly made product. The DNA containing the dye can be measured on a special instrument that provides real time information about what DNA containing the dye is present in the reaction and how much.
One question commonly asked is “what does a qPCR test of eDNA actually tell us”. qPCR results are basically a single snapshot in time of the site sampled. The signal produced by the eDNA during the qPCR test relates to the presence or absence of a specific species, and the approximate density of that species at a the location tested at the time the sample was taken. The detection of eDNA from a fish species does not provide information about the age or sex of individuals present at the time of sampling, and does not indicate whether the DNA came from a live organism or a recently dead one (for example a bait fish). Detection sensitivity is limited by how far away from the original source of the DNA the water was collected. The dispersal of eDNA in the environment is affected by factors like rapid water flow or wind. In addition, the rate at which eDNA breaks down is affected by environmental conditions like temperature and the local bacterial community. If the qPCR test signal does not give a positive signal, that may mean that the species being tested for is not present, or is there in such low abundance that the signal cannot be detected. A weak eDNA signal could represent a few cells from a non-resident fish that was transiently at the site tested and left behind some shed cells, or a fish that has recently died, or a fish that has just entered the site and has not yet deposited many cells in the water. A strong signal suggests the presence of a larger population of fish. Secondary testing of the same site later in time can help establish patterns of fish populations. Overall, qPCR is a very sensitive test that can be used to identify specific fish species even when they are present at very low numbers. For example, qPCR analysis of grass carp eDNA in a single water sample has picked up the presence of one grass carp in a 50 acre pond 10 feet deep under good conditions, although a lower sensitivity of one grass carp in a 2 acre pond has been seen when environmental conditions favor the rapid breakdown of shed cells.
What happens to the data collected?
The information generated by the qPCR analysis is incorporated into a fish species database identifying all locations tested, and indicating those sites that produced a positive test for any of the species being monitored. This information can help provide a clearer overview of the extent of invasive species throughout New York and will be very useful in framing potential responses. A map depicting all monitoring locations and the results obtained is publicly available. In addition, the qPCR results are returned to the teachers and students for classroom analysis and discussion.